
Look inside the largest spatial omics dataset in oncology
We are making a 60-patient sample from the larger MOSAIC dataset available to researchers. We call this MOSAIC-Window.
Benefits
Work with cutting edge modalities such as spatial omics and single-cell data
Run spatial omics proof-of-concepts before moving to larger projects
Harness spatial omics data for a greater chance of clinical success
New therapeutic targets for cancer indications
New patient subgroups
New diagnostic and prognostic biomarkers
New treatment response and resistance mechanisms


Patient samples
2,700
Patient samples
60
Cancer indications
NSCLC, Ovarian, Bladder, Mesothelioma, Glioblastoma, Breast, DLBCL, Head & Neck, Pancreatic, Colorectal
Cancer indications
Glioblastoma, Bladder, Ovarian, DLBCL, Mesothelioma
Data modalities
Spatial Transcriptomics, Single-Cell RNA-Seq, Bulk RNA-Seq, Clinical data, WES, H&E
Data modalities
Spatial Transcriptomics, Single-Cell RNA-Seq, Bulk RNA-Seq, Clinical data, WES, H&E
How to access MOSAIC Window data
Option 1 - Access via the EGA
The MOSAIC-Window dataset is available via the European Genome-Phenome Archive (EGA), a global network for archiving and sharing of data resulting from biomedical research projects.








Step 1
Researchers in academia and industry can apply to access the MOSAIC-Window dataset
Step 2
Applications are subject to approval by Owkin and the MOSAIC partners
Step 3
Run your research project using the data
Step 4
Resulting publications will cite MOSAIC and its partners
Option 2 - Explore via K Pro
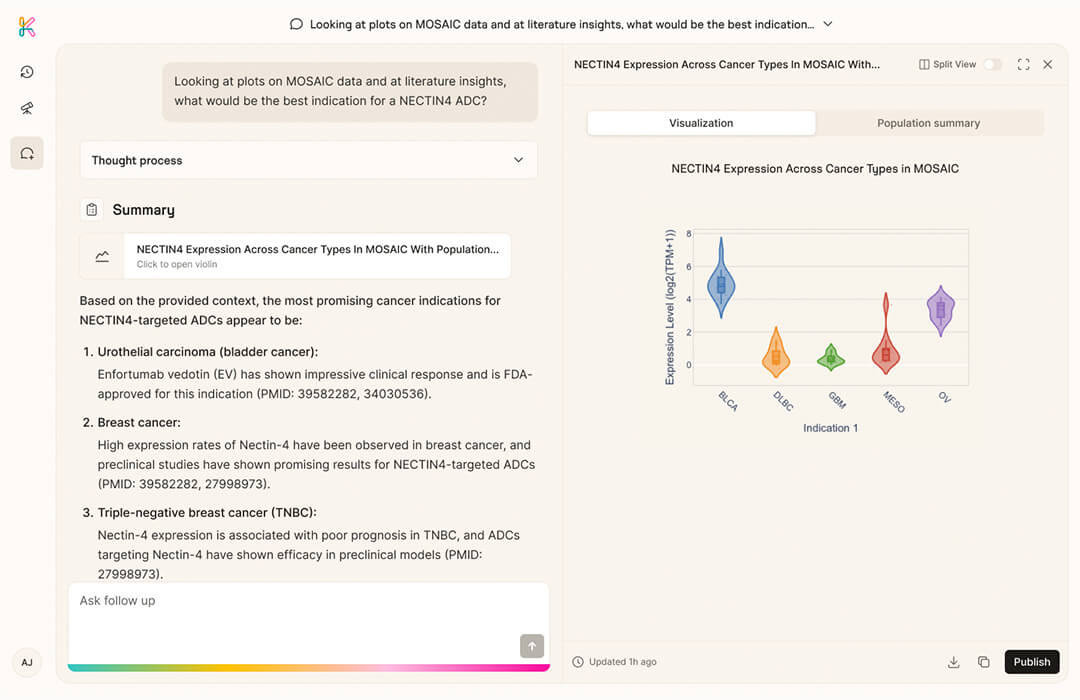

You can explore the MOSAIC-Window dataset via the free tier of our agentic AI co-pilot K Pro Free.
